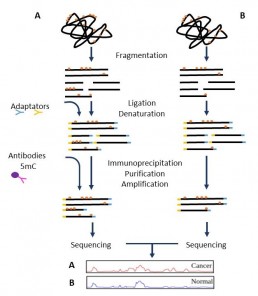
Mapping of DNA epigenetic marks: MeDIP
DNA modifications are epigenetic mechanisms. Among them is the methylation of cytosines (5mC) located in CpG sites or CpG islets close to the regions promoting transcription. The methylation state of these loci will suppress gene expression by disrupting the binding of transcription factors or altering the structure of the chromatin. The comparison of MeDIP(Methylated DNA Immunoprecipitation) profiles between 2 samples allows assessing the role of DNA methylation on certain disease conditions (cancer).
The analysis of the genome wide methylation status is achieved by sequencing MeDIP libraries. The DNA is fragmented by sonication, indexed with sequencing adaptors and then incubated with anti-5mC antibodies bound to magnetic beads.
Methylated complex (bead-antibody-fragments) are collected using a magnet, thus removing from the sample any unmethylated or weakly methylated molecules: the sample enriched with methylated sequences is ready to be analysed by sequencing.
For better reproducibility, it is recommended to use a robot to perform MeDIP.
PLATEFORMS TO CONTACT FOR “MeDIP” SEQUENCING PROJECTS
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq