A network of specialised and complementary platforms
The FRANCE GENOMIQUE infrastructure brings together the majority of sequencing and/or bioinformatics platforms in France:
- the national platforms of Genoscope and CNRGH in Evry, provide sequencing, genotyping and bioinformatics capacities to enable the completion of very large-scale projects,
- Sequencing platforms each with their own specific expertise and technologies as well as ad hoc bioinformatics tools
- the platforms associated with FRANCE GENOMIQUE,
- the CEA's TGCC where data storage and processing spaces are allocated to FRANCE GENOMIQUE partners.
- FRANCE GENOMIQUE has also recognised a number of platforms that are members of the IFB (Institut Français de Bioinformatique) as associated bioinformatics platforms, working in collaboration with the FRANCE GENOMIQUE sequencing platforms and the beneficiaries of these platforms.
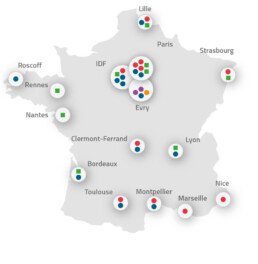
Fly over the name of the platform on the map and click to access the platform details.
- National sequencing platforms
- Sequencing platforms
- Associated sequencing platforms
- Associated bio-informatic platforms
- TGCC : Très Grand Centre de Calcul
- National sequencing plateforms
- Local sequencing plateforms
- Bio-informatic plateforms
- Associated plateforms
- TGCC : Très Grand Centre de Calcul
Very high throughput and 3rd generation sequencing instruments
FRANCE GENOMIQUE is equipped with the most efficient and innovative technologies dedicated to sequencing. The fleet is evolving very rapidly with very high throughput (NovaSeq 6000) and so-called 3rd generation sequencing instruments (unique molecule technology).
FRANCE GENOMIQUE is thus able to respond to all requests for sequencing projects
An high performance computation infrastructure
The CEA “Très Grand Centre de Calcul” (TGCC) is an infrastructure dedicated to high-performance computing, capable of hosting petaflop-scale supercomputers and designed on the basis of a data-driven architecture. Within the TGCC, the CCRT has an extension dedicated to the users of the FRANCE GENOMIQUE project.
This e-infrastructure for data storage and processing, implemented by the CEA/DIF teams, allows GENOMIC FRANCE users to benefit from a medium-term storage space (scale: scientific projects lasting several years) of several petabytes, connected to several thousand scalar computing cores by a high-performance interconnection. As the volumes of data to be stored and processed increase exponentially, it is also designed to be scalable, with the objective of meeting all the challenges of genomics in the future.
Equipment and capacity
The set up dedicated to FRANCE GENOMIQUE is composed of :
- 180 dual processor nodes (Intel Sandy Bridge E5-2680, 2.7 GHz, 8 cores) with 128 GB of memory per node, i.e a total of 2,880 cores (Bull),
- 2 Bullx S6410 very large memory systems with 2 TB of memory,
- 9 hybrid blades, equipped with nvidia Kepler GPUs.
Data hosting is achieved through the following storage configuration:
- Medium-term storage with a global file system of 5 PB, including 2 PB on disk (hierarchical storage system Lustre + IBM HPSS),
- Archive system for preliminary data.
Expertise and support
The CEA/DIF teams have developed internationally recognized expertise and competence both in the management of very large volumes of data (contribution to Opensource developments, management of EOFS, etc.) and in the definition and management of very large data centres. Assistance and support teams are at hand to help users make the most of the centre resources.
A dedicated application support team is provided by the national platforms (CEA) on behalf of FRANCE GENOMIQUE.
Main achievements
In order to characterize a set of 83 protein families without known functions and including some 60,000 sequences, Genoscope researchers conducted a modelling campaign on the CCRT Titane supercomputer. This phase, which would have required 280,000 hours of computation, could be performed in only 70 hours on 4,000 processors. From the results, the researchers created a catalogue of specific structural signatures for each of the families studied. This catalogue will provide biochemists with valuable information to discover new enzymatic activities.
The Genoscope has been using the TGCC/CCRT calculation resources for several years now, particularly via the DARI calls for projects. In this context, the TARA OCEANS project has benefited from more than 3.5 million hours of calculations to study the diversity of marine organisms. To do this, various sequence analysis tools have been ported to this infrastructure: BLAST, BLAT, InterProScan & CDDsearch. Specific codes have been designed and deployed to adapt these tools to the technical constraints of operating CCGT machines (massive data parallelization, execution control, error recovery, short unit jobs).
For further information
Web site : www-hpc.cea.fr/, www-ccrt.cea.fr
Plateform manager : Pierre Leca
CEA DAM-île de France
Bruyères-le-Châtel
91297 Arpajon Cedex
Contact: e-infrastructure@france-genomique.org

Illumina offers high and very high throughput sequencing.
After clonal amplification of short DNA fragments, sequencing by synthesis (SBS) begins: each base emits a unique fluorescence signal when added to the strand being synthesized. The detection of the signal at each incorporation determines the DNA sequence.
MGI’s DNA sequencing instruments utilize the core technology called DNBSEQTM.
DNBs (DNA nanoballs) are pumped with by the fluidics system and loaded onto a Patterned Array chip.
Sequencing primer is then added and hybridized to the adaptor region of the DNB. The sequencing reaction starts by pumping sequencing reagents containing fluorescently labeled dNTP probes and DNA polymerase. Images are taken after the fluorescently labeled probes on the DNB are excited with lasers. The images are then converted into a digital signal. This information is then used to determine the DNA sequence of the sample.

The PacBio sequencing platform is a long-read sequencing platform.
The core technology, single-molecule real-time (SMRT), can generate reads tens of kilobases in length. SMRT sequencing largely avoids sequence-specific biases in the NGS system, as most PCR amplification steps are not required in the library construction process.
Oxford Nanopore Technologies offers a technology for real-time DNA and RNA sequencing without synthesis and amplification, where the sequencing is carried out through a nanopore subjected to an electric field.
The ionic current differs according to the base A, T, G or C that blocks the nanopore. The identification of the sequence is done by measuring the evolution of the ionic current passing through the nanopore

The AVITI sequencer from Element Biosciences provides medium-scale sequencing at higher quality and significantly lower costs compared to Illumina sequencers.
While the sequencing chemistry is very different, the instrument is compatible with Illumina libraries. In contrast to the traditional sequencing-by-synthesis chemistry (SBS), the AVITI employs a sequencing-by-binding chemistry (SBB) that requires the binding of a multivalent fluorescent polymerase substrate by avidity. The use of these so-called Avidites increases specificity and reduces the fluorescent chemistry costs by an order of magnitude. With this technology, the DNA synthesis steps can be carried out with unlabeled nucleotides.

The company 10x Genomics has developed a machine that partially solves the hurdles of short-reads synthetic sequencing (SBS): Chromium.
This system uses an emulsion PCR method. The purpose of creating the emulsion is to encapsulate in a drop of liquid containing all reagents needed, few high molecular weight DNA molecules in the case of a “long synthetic reads sequencing”, or a cell in the case of “single cell sequencing”.
This method allows short reads assemblies (Illumina) to be made via a single barcoding system, facilitating phasing analysis and characterization of chromosome structures. It also allows to study the transcriptome by RNAseq of several thousand unique cells in parallel. After preparing the libraries, the sequencing is performed on Illumina machines.
More recently, 10X Genomics has developed a solution that integrates single-cell transcript detection with high-resolution imaging for the decoding and analysis of spatial transcriptomics data.