“Mate Pair” Sequencing
The preparation of “mate pair” libraries is designed to allow classical “paired-end” sequencing of both ends of a fragment with an original size of several kilobases.
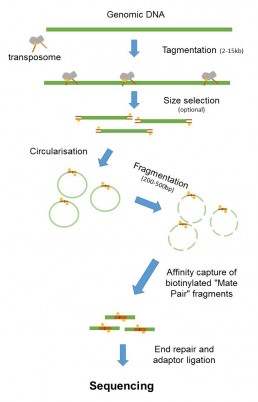
The figure shows the workflow for “mate-pair” library preparation for Illumina sequencing. Since the beginning of 2013, this preparation has been based on Nextera technology. The enzyme tagmentase fragments the DNA to sizes between 2 and 15 kb and binds adapters at the breaking point. Fragment size can be selected by 0.7% agarose gel migration or pulse field. DNA inserts are then circularised to bring the ends close together. At the junction point, a biotin is added and the circularised constructions are mechanically sheared into fragments of 200 to 700pb. The segments containing the ends are then recovered using a magnetic streptavidine bead selection. The fragments selected in this way are then submitted to a standard library preparation.
Platforms to contact for “mate pair” sequencing projects