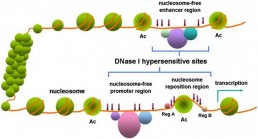
Mapping of chromatin accessibility sites: DNase seq
Mapping regions of relatively open chromatin is valuable for identifying regulatory elements, including promoters and enhancers. DNase-seq combines cleavage of Dnase1 hypersensivitive sites (see fig) and high throughput sequencing allowing the identification of most active regulatory regions for a specific cell type.
DNase-seq protocol starts by a high quality nuclei isolation which is cell line depend. Nuclei are then incubated with DNase 1. The DNA cleavage by the enzyme must be optimised to avoid sporadic or overdigestion of the chromatin. After RNA and proteins degradation using RNases and Proteinase K, respectively, the DNA is purified and size-selected for class of factors to study (typically 50–100 bp for transcription factors and 130–160 bp for nucleosomes. Isolated fragments are the used as template for library construction and sequencing (Chen et al. 2018)
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- DNA binding sites map : CUT & RUN
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq